Transfer of plasmid DNA into bacteria. How bacteria are selected. Protein production and purification.
[Source: Khan Academy]
Key points:
- Bacteria can take up foreign DNA in a process called transformation.
- Transformation is a key step in DNA cloning. It occurs after restriction digest and ligation and transfers newly made plasmids to bacteria.
- After transformation, bacteria are selected on antibiotic plates. Bacteria with a plasmid are antibiotic-resistant, and each one will form a colony.
- Colonies with the right plasmid can be grown to make large cultures of identical bacteria, which are used to produce plasmid or make protein.
The big picture: DNA cloning
Transformation and selection of bacteria are key steps in DNA cloning. DNA cloning is the process of making many copies of a specific piece of DNA, such as a gene. The copies are often made in bacteria.
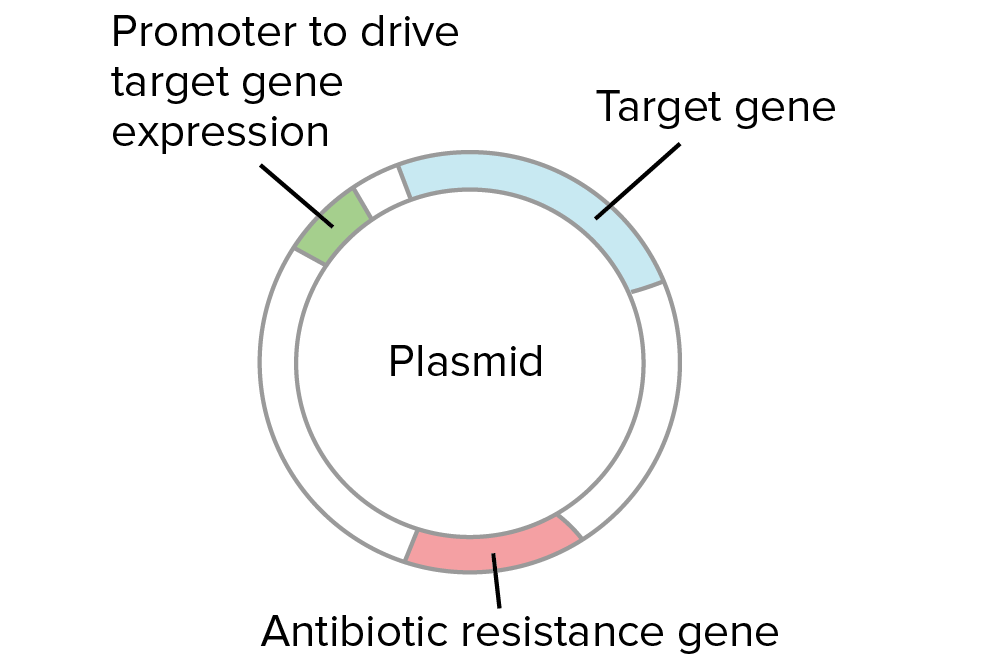
In a typical cloning experiment, researchers first insert a piece of DNA, such as a gene, into a circular piece of DNA called a plasmid. This step uses restriction enzymes and DNA ligase and is called a ligation.
After a ligation, the next step is to transfer the DNA into bacteria in a process called transformation. Then, we can use antibiotic selection and DNA analysis methods to identify bacteria that contain the plasmid we’re looking for.
Steps of bacterial transformation and selection
Here is a typical procedure for transforming and selecting bacteria:
- Specially prepared bacteria are mixed with DNA (e.g., from a ligation).
- The bacteria are given a heat shock, which causes some of them to take up a plasmid.
- Plasmids used in cloning contain an antibiotic resistance gene. Thus, all of the bacteria are placed on an antibiotic plate to select for ones that took up a plasmid.
- Bacteria without a plasmid die. Each bacterium with a plasmid gives rise to a cluster of identical, plasmid-containing bacteria called a colony.
- Several colonies are checked to identify one with the right plasmid (e.g., by PCR or restriction digest).
- A colony containing the right plasmid is grown in bulk and used for plasmid or protein production.
Why do we need to check colonies?
The bacteria that make colonies should all contain a plasmid (which provides antibiotic resistance). However, it’s not necessarily the case that all of the plasmid-containing colonies will have the same plasmid.
How does that work? When we cut and paste DNA, it's often possible for side products to form, in addition to the plasmid we intend to build. For instance, when we try to insert a gene into a plasmid using a particular restriction enzyme, we may get some cases where the plasmid closes back up (without taking in the gene), and other cases where the gene goes in backwards.
Why does it matter if a gene goes into a plasmid backwards? In some cases, it doesn't. However, if we want to express the gene in bacteria to make a protein, the gene must point in the right direction relative to the promoter, or control sequence that drives gene expression. If the gene were backwards, the wrong strand of DNA would be transcribed and no protein would be made.
Because of these possibilities, it's important to collect plasmid DNA from each colony and check to see if it matches the plasmid we were trying to build. Restriction digests, PCR, and DNA sequencing are commonly used to analyze plasmid DNA from bacterial colonies.
Protein production in bacteria
Suppose that we identify a colony with a "good" plasmid. What happens next? What's the point of all that transforming, selecting, and analyzing?
Possibility 1: Bacteria = plasmid factories
In some cases, bacteria are simply used as "plasmid factories," making lots of plasmid DNA. The plasmid DNA might be used in further DNA cloning steps (e.g., to build more complex plasmids) or in various types of experiments.
In some cases, plasmids are directly used for practical purposes. For instance, plasmids were used to deliver a human gene to lung tissue in a recent gene therapy clinical trial for patients with the genetic disorder cystic fibrosis.
Possibility 2: Bacteria = protein factories
In other cases, bacteria may be used as protein factories. If a plasmid contains the right control sequences, bacteria can be induced to express the gene it contains when a chemical signal is added. Expression of the gene leads to production of mRNA, which is translated into protein. The bacteria can then be lysed (split open) to release the protein.
Bacteria contain many proteins and macromolecules. Because of this, the newly made protein needs to be purified (separated from the other proteins and macromolecules) before it can be used. There are a variety of different techniques used for protein purification.
In one technique called affinity chromatography, a mixture of molecules extracted from the lysed bacteria is poured through a column, or a cylinder packed with beads. The beads are coated with an antibody, an immune system protein that binds specifically to a target molecule.
The antibody in the column is designed to bind to our protein of interest, and not to any other molecules in the mixture. Thus, the protein of interest is trapped in the column, while the other molecules are washed away. In the final step, the protein of interest is released from the column and collected for use.
[Source: Khan Academy]